
In our group, we tackle fundamental questions of life, such as gene expression and DNA replication in eukaryotes, as well as the chromatin structural changes that regulate these processes. Our unique approach combines world-leading single-molecule imaging techniques with large-scale molecular simulations using supercomputers. By integrating experiments, simulations, and machine learning, we aim to elucidate the molecular-level dynamics underlying essential chromatin-based processes such as transcription, replication, and repair, and to contribute to a deeper understanding of epigenetics and cell fate regulation. We are always welcoming new members. If you are passionate about understanding life at the molecular level or wish to acquire cutting-edge experimental and computational skills, we encourage you to join us — regardless of your background in life sciences, physics, chemistry, or information science. If you are interested, please feel free to contact us by email (terakawa at biophys.kyoto-u.ac.jp).
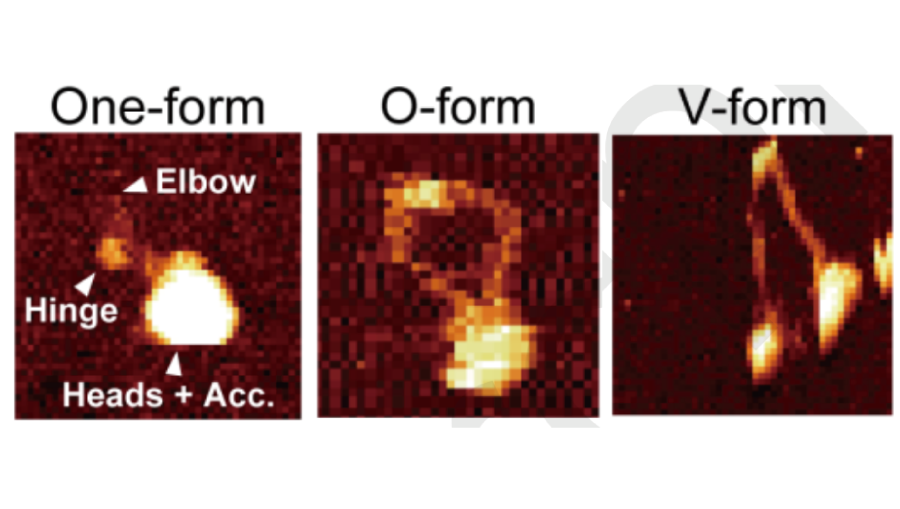
Condensin dynamics
We analyzed the condensin dynamics associated with ATP binding using solution AFM and molecular simulations. ATP-induced head engagement is coupled with hinge opening. This work was conducted in collaboration with the Kodera group at Kanazawa University.

Cohesin DNA Binding
Using coarse-grained simulations, we identified DNA binding sites in the human cohesin. DNA binding patches were found on SMC1, SMC3, STAG1, and NIPBL, and were shown to cooperatively bend DNA. Furthermore, STAG1 and NIPBL were revealed to support DNA loop extrusion.
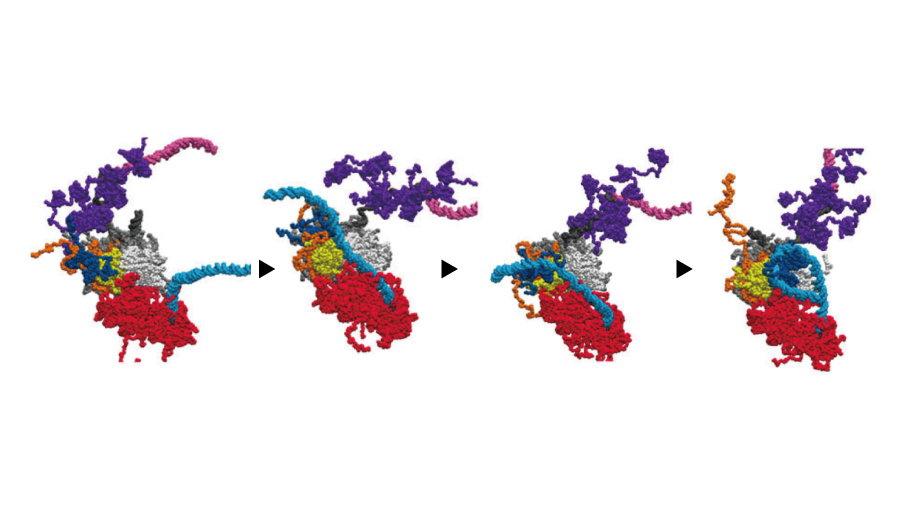
Histone Recycled
We visualized parental histone H3/H4 recycling at the DNA replication fork using coarse-grained molecular dynamics simulations. Histones were re-deposited via Cdc45-dependent and -independent pathways, with RPA and Pol ε binding influencing strand selection and recycling sites.
MutS Corkscrewing
Using coarse-grained molecular dynamics simulations, we visualized the dynamics of MutS, a DNA base pair mismatch recognition protein, sliding along DNA while rotating around DNA. As a result, we found that the diffusion rate of MutS changed according to the DNA bending.

Lane Switch on Nucleosome
By verifying the coarse-grained molecular dynamics simulation results with biochemical experiments and nanopore sequencing, we revealed that DNA translocases push nucleosomes along DNA. We proposed the "Lane Switching" as the molecular mechanism.

Condensin is a Motor
Single-molecule fluorescence imaging using a DNA curtain device revealed that condensin, involved in the chromatin structural formation, is a molecular motor. The results were published in Science and featured on the cover. Now, we are studying its molecular mechanism.
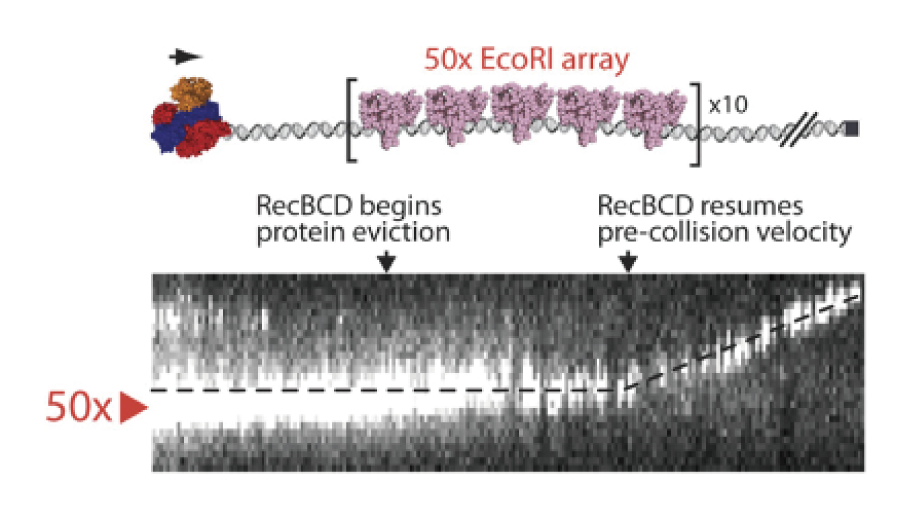
RecBCD Sequential Eviction
Using single-molecule fluorescence imaging, we observed the RecBCD molecular motor dynamics as it encountered densely bound protein arrays on DNA. The results revealed that RecBCD sequentially evicts individual proteins, allowing it to efficiently translocate along crowded DNA.

RESPAC: CG Electrostatics
We developed the RESPAC method to refine charge assignment in coarse-grained protein models based on atomic-level electrostatic potential. The method significantly improved the reproduction of electrostatic potential compared to conventional integer-charge models.
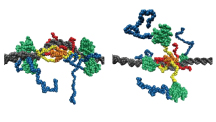
p53 Dancing on DNA
Using coarse-grained molecular dynamics simulations, we visualized p53, a tumor-suppressive transcription factor, sliding on DNA. As a result, it was suggested that the DNA-binding region searches for the recognition site by "dancing" while diffusion on the DNA.
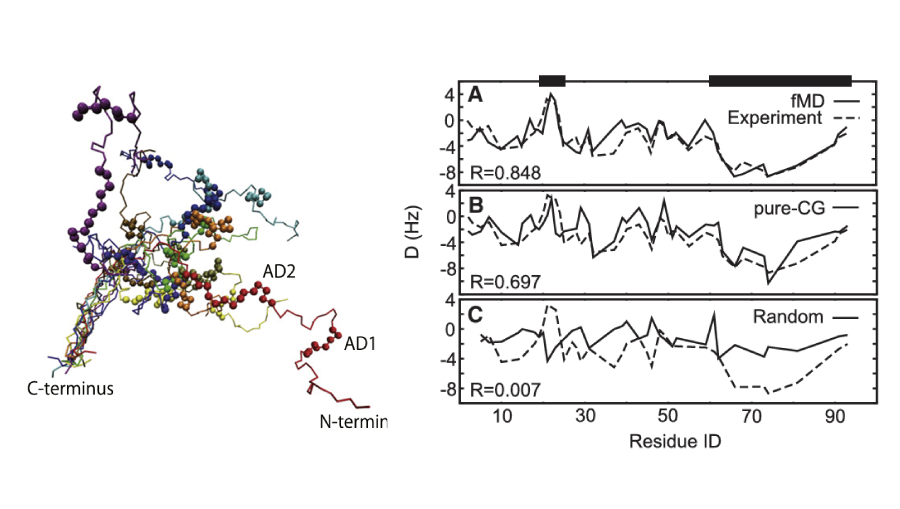
CG for disorder
We developed a multiscale approach that integrates all-atom molecular dynamics simulations of fragments with coarse-grained modeling, enabling high-accuracy reproduction of structural ensembles, which showed good agreement with experimental data.
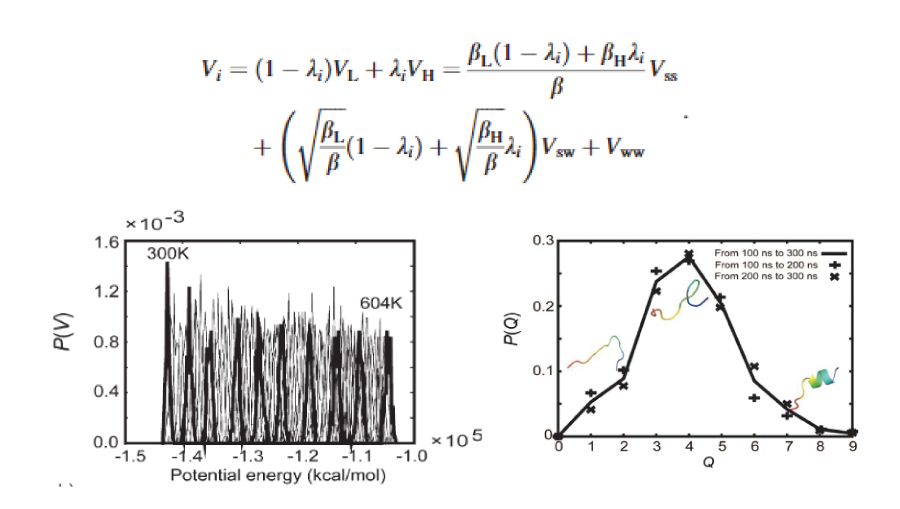
Solute Tempering
We demonstrated that a variant of the REST method can be easily implemented in GROMACS by rescaling force-field parameters. Then, we confirmed that this approach achieves comparable sampling efficiency to conventional REM while requiring significantly fewer replicas.









